 (Source: Holtmaat Lab at University of Geneva, Switzerland)
(Source: Holtmaat Lab at University of Geneva, Switzerland) We design novel distributed algorithms and build an open-source software implementation of a Web-based system for collaborative automated and manual tracing of neurites from light-microscopy stacks of images. This is a joint project with the Neurogeometry Lab of Prof. Armen Stepanyants. It builds on the success of NCTracer, turning it from a standalone software that runs on a single machine to a collaborative and scalable system that makes the power of distributed (cloud) computing available to neuroscientists through a standard Web browser.
NCTracer Web is designed to support collaborative tracing and analysis of multi-terabyte-size brain images we receive from national and international collaborators. In addition to the concrete challenges posed by brain-connectivity analysis, we are designing distributed algorithms that also apply to other domains, e.g., social-network analysis and SQL database queries.
 (Source: Holtmaat Lab at University of Geneva, Switzerland)
(Source: Holtmaat Lab at University of Geneva, Switzerland)
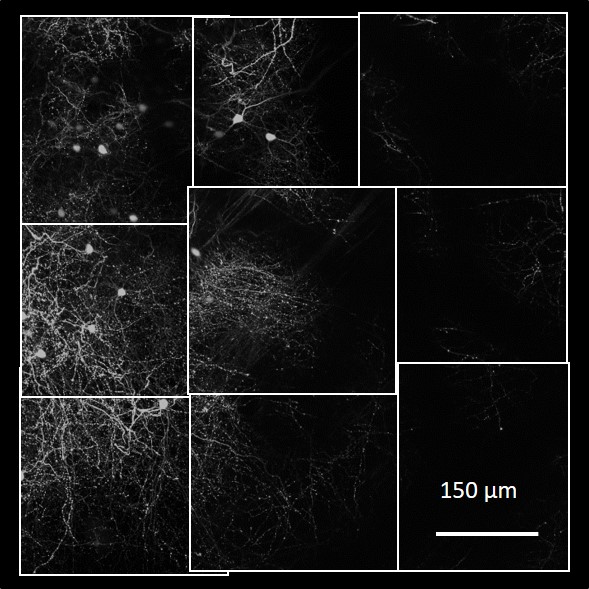
Light-microscopic imaging of a mouse brain produces about 20,000 3D image stacks, each typically 1024x1024x312 voxels and covering about 0.26x0.26x0.8 micrometers3, for a total of about 10 terabytes of TIFF data. These image stacks overlap, but their microscope-recorded positions and orientation are not sufficiently precise. For scientific analysis, the following major processing steps are required:
Registration: Turn the individual stacks into a coherent 3D image of the entire brain, ensuring correct connections across stack boundaries.
Automatic and manual tracing: Convert the image into a graph that represents axons and dendrites as nodes and edges. This is challenging in dense regions where many connections cross. Since manual tracing is too slow, we follow a combined approach of scalable automatic tracing, followed by manual tracing to correct potential errors.
Graph analytics: Once the image is represented as a graph, we can explore its structure, e.g., find common patterns or determine the region of influence of a neuron.
 (Source: Stepanyants Lab at Northeastern University)
(Source: Stepanyants Lab at Northeastern University)
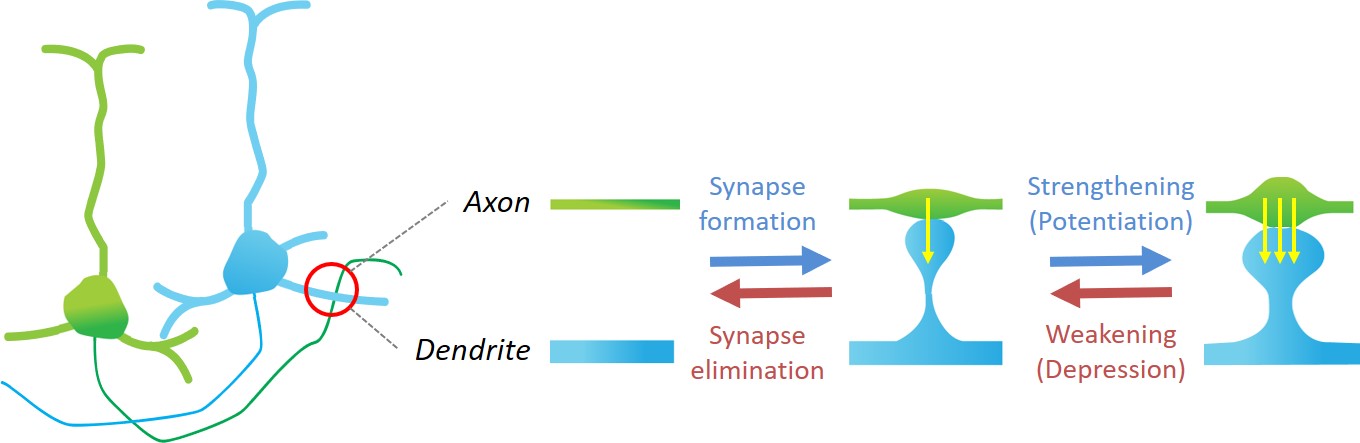
In addition to static full-brain images, we also have access to time-series data taken periodically over a large time span in the same animal. These images cover only a fraction of the brain and only the outer 1mm of it, but they enable studying dynamics of how synapses, which connect an axon of a neuron to the dendrite of another, form or disappear over time.
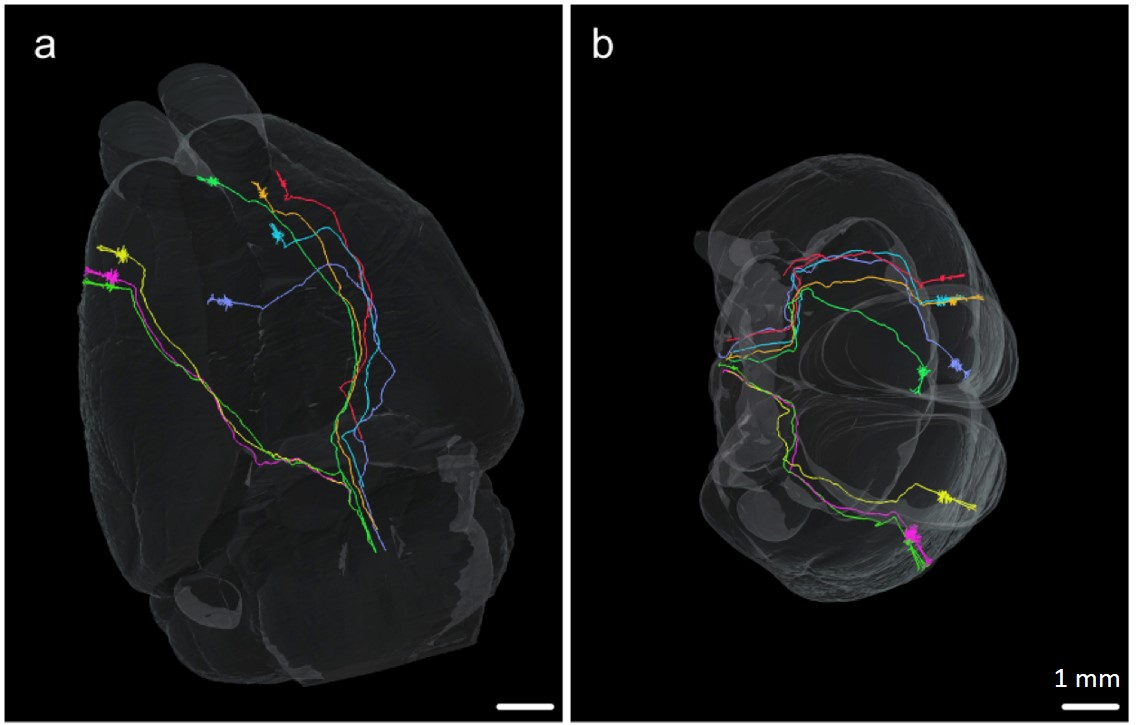 (Source: Zheng T. et al. Visualization of brain circuits using two-photon
fluorescence micro-optical sectioning tomography, Optics Express, 21(8):
9839-9850, 2013)
(Source: Zheng T. et al. Visualization of brain circuits using two-photon
fluorescence micro-optical sectioning tomography, Optics Express, 21(8):
9839-9850, 2013)
Motivated by the brain-data analysis application and by potential applications to other domains, we are studying algorithmic challenges such as:
Image alignment in space and time: Align 20,000 3D image stacks in space and do this efficiently in a distributed system with tens or hundreds of machines. Align time-series data in space and time. What are general principles that apply to these two and other image-alignment problems so that new alignment challenges require only minor customization instead of having to design a completely new approach from scratch?
Top-k pattern search in graphs: Identify common structural patterns and search for specific graph patterns. When the number of result patterns is very large, we want to quickly return the most relevant patterns, then keep returning the next-most-relevant pattern instances until the user is satisfied.
Distributed joins: Joins are crucial for pattern search and correlation finding. Design effective distributed algorithms for common types of joins, including equi-join and band-join, but also the general theta-join. Can we prove near-optimality properties, e.g., for the largest amount of load assigned to any machine in the system?
Evolving graphs: What is the best way to store, manage, and query evolving graphs?
We have a variety of strong initial results, including provably near-optimal algorithms for distributed theta-joins and equi-joins. The latter (as of late 2019) represent the best known distributed equi-join algorithms---in terms of running time---for skewed input. However, many open research problems are waiting for curious and motivated PhD students and post docs. And we also have projects for undergraduate and Masters students.
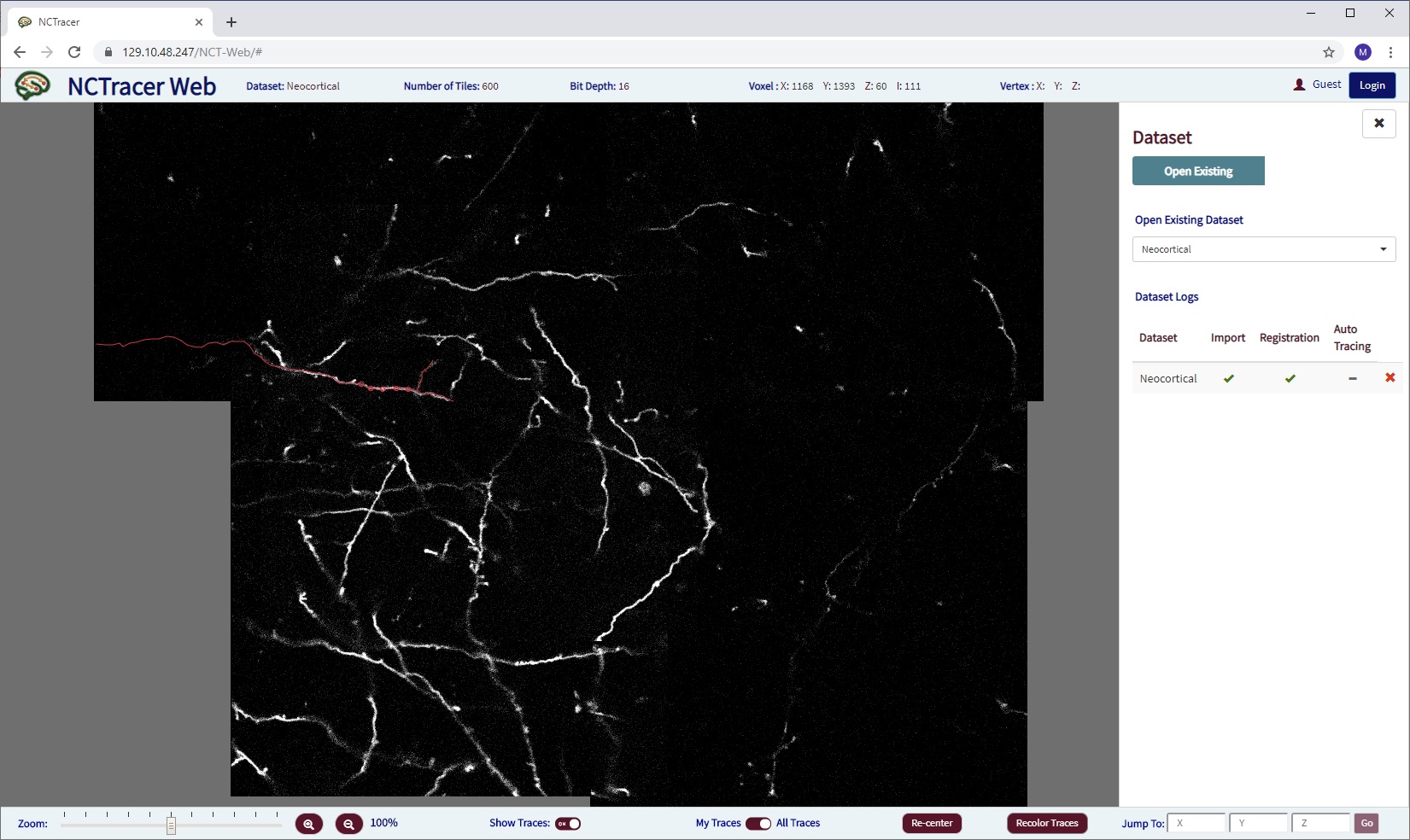
The current version of our NCTracer Web system demo is running here. (We recommend using the Chrome browser.) It is under development, therefore may be running at a different address. Here is how it looked as of late September 2019:
Rundong Li, Wolfgang Gatterbauer, Mirek Riedewald. Near-optimal distributed band-joins through recursive partitioning. To appear in Proc. ACM SIGMOD Int. Conf. on Managament of Data, 2020 [preprint]
Nikolaos Tziavelis, Wolfgang Gatterbauer, Mirek Riedewald. Optimal join algorithms meet top-k. To appear in Proc. ACM SIGMOD Int. Conf. on Managament of Data, 2020 (tutorial) [preprint]
R. Li, N. Mi, M. Riedewald, Y. Sun, Y. Yao. Abstract Cost Models for Distributed Data-Intensive Computations. Distributed and Parallel Databases, 37(3): 411-439, Springer, 2019 [preprint]
R. Li, M. Riedewald, X. Deng. Submodularity of Distributed Join Computation. In Proc. ACM SIGMOD Int. Conf. on Managament of Data, pages 1237-1252, 2018 [preprint]
X. Yang, D. Ajwani, W. Gatterbauer, P. K. Nicholson, M. Riedewald, and A. Sala. Any-k: Anytime Top-k Tree Pattern Retrieval in Labeled Graphs. In Proc. Int. Conf. on World Wide Web (WWW), pages 489-498, 2018 [extended version]
R. Li, N. Mi, M. Riedewald, Y. Sun, Y. Yao. A Case for Abstract Cost Models for Distributed Execution of Analytics Operators. In Proc. Int. Conf. on Big Data Analytics and Knowledge Discovery (, pages 149-163, 2017 (invited to special issue of Distributed and Parallel Databases presenting the Best Papers of DaWaK) [paper]
A. Okcan and M. Riedewald. Processing Theta-Joins using MapReduce. In Proc. ACM SIGMOD Int. Conf. on Managament of Data, pages 949-960, 2011
Xinyan Deng (MS student, first job after graduation: Microsoft)
Rundong Li (PhD student, first job after graduation: Google)
Mirek Riedewald
Seyed Kahaki
(postdoc)
Armen
Stepanyants
This project would not have been possible without the creativity and hard work of the following students, most of them MS students, who contributed to the code (names shown in temporal order starting with the most recent contributors): Xuying ``Jeanne'' Shao, Shubham Muttepawar, Vivin Wilson, Ding Jin, Han Fu, Anubhuti Vyas, Chuhan Liu, Yogesh Gupta, Paurav Patel, Sarita Joshi, Yogiraj Awati, Dhavalkumar Patel, Ankur Shankar Shanbhag, and Joe Sackett.
Prof. Riedewald and his students' work on this project was supported by the National Science Foundation (NSF) under award no. 1017793, the National Institutes of Health (NIH) under award no. R01 NS091421, and by PwC (PricewaterhouseCoopers). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of NSF, NIH, or PwC.


