CSG339 Information Retrieval
Project 1
Courtesy to James Allan
http://www-ciir.cs.umass.edu/~allan/
Created: Wed 20 Sep 2006
Last modified:
Assigned: Thu 21 Sep 2006
Due: Thu 19 Oct 2006
Project 1
Q&A
Lemur
details
Sample:q57_d3_laplace
Sample AP numbers per query
This project requires that you use a server to
build variations on a simple search engine. For each variation you will run
and evaluate a number of queries.
Building an IR system
This project requires
that you construct a search engine that can run queries on a corpus and
return a ranked list of documents. Rather than having you build a system
from the ground up, we are providing a Web interface to an existing search
engine. That interface provides you with access to corpus statistics,
document information, inverted lists, and so on. You will access it using
the protocol, so can implement your project in any language that provides you
with Web access, and can run it on any computer that has access to the Web.
Computers in the CCISS department's will work, but you are more than welcome
to use your own computer.
One of the nice things about the CGI interface is that you can get an idea of
what it does using any browser. For example, the following CGI
command:
http://fiji4.ccs.neu.edu/~zerg/lemurcgi/lemur.cgi?v=encryption
will get you a list of the documents that contain the word "encryption". Go
ahead and click on the link to see the output (in a separate window). It
lists the number of times the term occurs in the collection (ctf), the total
number of documents that contain it (df) and then, for each of the documents
containing the term, it lists the documents id, it's length (in bytes), and
the number of times the term occurs in that document.
The format is easy for a human to read and should be easy for you to write a
parser for. However, to make it even simpler, the interface provides a
"program" version of its interface that strips out all of the easy-to-read
stuff and gives you a list of numbers that you have to decipher on your own
(mostly it strips the column headings). Here is a variation of the link
above that does that. Click on it to see the difference.
http://fiji4.ccs.neu.edu/~zerg/lemurcgi/lemur.cgi?g=p&v=encryption
From class, it should be pretty straightforward to imagine how to create a
system that reads in a query, gets the inverted lists for all words in the
query, and then comes up with a ranked list. You may need other
information, and the interface makes everything accessible (we hope).
Hema's wiki page contains a rundown of some of the commands, and there is a
help command, too:
http://fiji4.ccs.neu.edu/~vip/lemurcgi/lemur.cgi?h=
Play around with the engine to ensure it makes sense.
Three very important tips. First, there is some chance that the servers for
this project will change, depending on load. Write all of your code such that
the name of the server ("fiji4.ccs.neu.edu" right now) is a variable or
constant that can easily be changed.
Second, in order to get efficient processing out of this server, you should
string as many commands together as you can. For example, if you want to
get term stemming, stopping, and statistics information for three terms, ask
for them all at once as in:
http://fiji4.ccs.neu.edu/~zerg/lemurcgi/lemur.cgi?c=encryption&c=star&c=retrieval
(If you use the program mode version of the same command, the Lemur images are dropped.) This is because there is
some overhead getting the process started, and putting commands together
amortizes that over several requests. It works to do them
separately, but it might take longer.
And third, some things will be inefficient this way, no matter what. You
may find it useful to use the file listing stem classes and the file that maps between
internal and external docidsrather than using calls to the server.
The databases
We have set up a database for you to use for this
assignment. They are taken from the AP89 subset of the TREC corpora and
consist of 84,678 news stories from the Associated Press in 1989 (here is
more information about the corpora if you're curious). In fact, we've set up four versions of
the database: all four combinations of using or not using each of stemming
and stopping. The collections contain the same documents, but the inverted
lists will obviously be different. The "d=?" command will provide you with
useful information about the databases.
The stop list that we used (for the two versions with stopping) is the
inquery stop list and includes 418 words. You can download a list of those words
here, since you will probably find it useful for your
own query processing. Note that the "c=term" command (and "t=term") also
shows the stopword status of a term (though be careful that you only pay
attention ot the stopword status of a word on a database that uses
stopping).
When stemming was applied, we used the kstem stemming algorithm. If you are
running on an unstemmed collection, use the "c=term" command to get term
statistics about the term. It will return stemming information, but you
should ignore it. If you are running on a stemmed collection, the "t=term"
command is also useful. Both "c=" and "t=" tell you the stemming and
stopping status of the word; however, the former gives you term statistics
for the original word rather than the stem and the latter gives you
term statistics for the stem rather than the original word. Having
both commands available can be useful because of the way Lemur does stemming:
it does not stem capitalized words that are not at the start of a sentence.
As a result, stemmed collections will have american converted to
america but American will not be changed. Whether this
behavior is "correct" or not is a matter of opinion; you just have to cope
with it. (By and large you will be safe assuming that stemming always
happens.) Here is a file that lists all stem classes appearing in this
database (the format is "root | variant1
variant2 ..."). It actually includes more words than are in AP89, but
everything in AP89 definitely appears.
The systems you build
Here is a set of 25 queries taken
from TREC in the format "querynum. query". These
queries have been judged by NIST against AP89 (using the pooling method). To
help you with your analysis below, the queries are sorted in order of the
number of relevant documents in the collection: the first query has 393
relevant documents and the last has only one (the complete distribution is
also available). Your assignment is to run those queries against the
-provided collection, return ranked lists in a particular format, and then
submit the ranked lists for evaluation.
Implement the following variations of a retrieval system:
Vector space
model, terms weighted by Okapi tf only (see note), inner product
similarity between vectors. Note that you currently do not have
sufficient information to calculate document length vector easily, so
pure cosine similarity isn't used.
For Okapi tf on documents use OKTF=tf/(tf + 0.5 +
1.5*doclen/avgdoclen).
On queries, Okapi tf can be computed in the same way, but use length of
the query instead of doclen.
Vector space
model, terms weighted by Okapi tf (see note) times an IDF value, inner
product similarity between vectors. The database information command
provides collection-wide statistics that you need.
You will have to use for the weights OKTF x IDF where OKTF is same as for system1
above.
Language modeling,
maximum likelihood estimates with LaPlace smoothing only, query
likelihood. The database information command provides collection-wide
statistics that you need.
If you use multinomial model, for every document, only the
probabilities associated with terms in the query must be estimated
because the others are missing from the query-likelihood formula (consult
slides on pages
13-19).
For model estimation use maximum-likelihood and LaPlace smoothing. Use
formula (for term i)
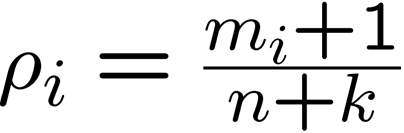
where
m=freq count (tf) , n=number of terms in document (doc length) , k=number
of unique terms in corpus.
Language modeling, Jelinek-Mercer smoothing
using the corpus, 0.8 of the weight attached to the background
probability, query likelihood. The database information command
provides collection-wide statistics that you need.
The formula for Jelinek-Mercer smoothing
is
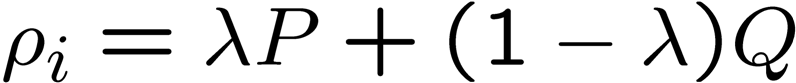
where P is the estimated
probability from document (max likelihood = m_i/n) and
Q is the estimated
probability from corpus (bkground probab = cf / terms in the
corpus).
- Language modeling, Dirichlet smoothing
- Language modeling, Whitten Bell smoothing
Note that if you look
up the "true" Okapi function, you find two things: (1) there are several
definitions of the "true" Okapi tf function and (2) it's got a few parameters
that you'd need to set. Depending on the form, if you set k1=1 and b=1.5,
or possibly set c=1, k1=2, and b=0.75, you'll end up getting this simplified
and fairly common version: tf / (tf + 0.5 + 1.5*doclen/avgdoclen).
Run all 25 queries and return the top 1,000 ranked documents for every query
(at most, less than 1000 because you should not include documents that got
score 0), putting that all into one file. So each file will have at most
25*1000 = 25,000 lines in it. The lines in the file must have the following
format:
query-number Q0 document-id
rank score Exp
where query-number is the number of the
query (the number between 51 to 100, for this project), document-id is
the external ID for the retrieved document (here is a map between internal and external IDs , rank is
the position in the ranked list of this document (1 is the best; 1000 is the
worst; do not include ties), and score is the score that you system
creates for that document against that query. Scores should descend while
rank increases. "Q0" (Q zero) and "Exp" are constants that are used by some
evaluation software. The overall file should be sorted by ascending rank
(so descending score) within ascending query-number
(remember that the queries are not in sorted order in the file).
Run all six models against all four database variations. That means you
will generate 6*4=24 files, for a total of at most 600,000 lines of ouput.
To evaluate a single run (i.e., a single file containing 25,000 lines or
less), you first need to download trec_eval (also see trec_eval_aprp) and the qrel_file which contains
relevant judgments; then run
trec_eval [-q] qrel_file results_file
(-q shows evaluations on all queries; without it you get only the averages).
It provides a pile of statistics about how well the uploaded file did for
those queries, including average precision, precision at various recall
cut-offs, and so on. You will need some of those statistics for this
project's report, and you may find others useful.
Using a simple tf-idf system without anything fancy, our baseline system
returns about 0.2 mean average precision. If your system is dramatically
under-performing that, you probably have a bug.
What to hand in
Provide a short description of what you did and
some analysis of what you learned. This writeup should include at least the
following information:
Uninterpolated
mean average precision numbers for all 16 runs.
Precision at 10
documents retrieved for all 16 runs.
An analysis of the benefits or disadvantages
of stemming, stopping, IDF, and different smoothing techniques.
Feel free to try other runs to better explore the
issues (how does Okapi compare to raw tf, how about that "augmented tf" thing
that was mentioned in class in regard to Smart, what happens if IDF is used
differently, what impact do different values of lamba have on smoothing,
etc.).
Hand in a printed copy of the report. The report should not be much longer
than a dozen pages (if that long). In particular, do not include trec_eval
output directly; instead, select the interesting information and put it in a
nicely formatted table.
Grading
This project is worth a possible 250 points. To
get about 200 points, you would do the bare minimum described above.
Additional points will be awarded (up to a maximum of 250) for doing things
such as:
Extra runs,
particularly interesting runs that do more than vary parameters, got
15-20 points. The more the better, though a small number of really
interesting runs will be worth more than a large number of
nearly-identical runs.
Graphs of the
recall/precision numbers are worth up to 15 points since they take some
time and present a much nicer illustration of the effectiveness,
particularly when multiple plots are combined on one graph for
comparison.
Analysis beyond the minimum required could
result in up to 25 points, though that would be an extreme. Good
analysis looks at something that was intriguing and investigates it in
more detail. It might require doing some additional runs (which would
give points for extra runs, too!). Examples include looking at
query-by-query performance to explore what makes some queries fail, or
playing with some query processing to get rid of stop structure
("documents will") or to otherwise process the queries.
![]()
![]()